
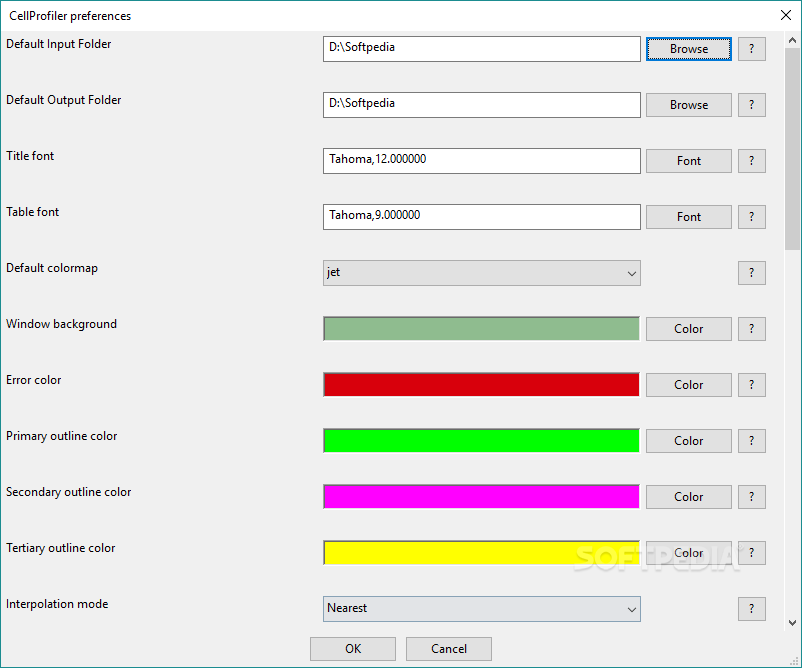
- Cellprofiler unmix color install#
- Cellprofiler unmix color update#
- Cellprofiler unmix color download#
Once the update is completed, click the Advanced mode menu and select Manage Update Sites and select Stowers Plugins from the menu. If you run FIJI, you can run the updater through Help>Update FIJI. On windows, this is typically c:\Program Files\ImageJ\Plugins. In regular ImageJ, you simply need to copy them to your ImageJ plugins folder.
Cellprofiler unmix color install#
You can install them by downloading them from http://zipped_plugins.html. Though this course will not necessarily rely heavily on my plugins, there are a few chapters that utilize these plugins. FIJI tries to make plugin installation simple by providing a large number of plugins preinstalled and working out dependencies. Plugin installation is another thing that is both an annoyance and an advantage in ImageJ. On almost all versions of ImageJ, the memory can be increased or decreased by running Edit>Options>Memory & Threads. That means that all commands to increase or decrease memory require you to restart ImageJ. Thirdly, Java requires you to allocate memory when you start ImageJ.
Cellprofiler unmix color download#
If you download ImageJ from the ImageJ or Fiji websites, you can specify which operating system you have and it will download the correct java along with it. Secondly, even if you have a 64-bit machine, your computer may or may not have 64-bit Java installed. 64-bit systems do not have this limitation and are therefore limited by the amount of RAM the computer has. If nothing is shown, the system is likely 32-bit. If you don't know whether your computer is 32-bit or not, right click on My Computer and go to the Properties menu and look for 32-bit or 64-bit somewhere on the resulting page. Firstly, Java limits 32-bit computers to around 1.5 GB of memory. A couple of important points about memory. ImageJ is written in the Java programming language, so limitations in Java apply to ImageJ.
This frustrates some users, but I feel that it is better to understand the memory limitations of a particular task rather than lock up the computer at some unknown point during analysis. ImageJ manages this process in a relatively transparent fashion. This is repeated repetitions times.Whether you like it or not, managing computer memory is an important part of all scientific image processing tasks. The algorithm shuffles one of the channels by dividing it into blocks of blockSizes pixels,Īnd randomly permuting these blocks. The methods listed below to quantify colocalization. Value to colocalization estimates, but does not quantify the amount of colocalization. Shuffling one of the channels, there no longer exists correlation. This test verifies whether there is colocalization in the image pair by comparing the correlationīetween the two channels to that of a randomly shuffled version of the channels. Stain color triplets taken from CellProfiler,ĭip::dfloat dip:: CostesSignificanceTest( dip::Image const& channel1,Ĭomputes Costes’ test of significance of true colocalization.Johnston, “Quantification of histochemical staining by color deconvolution”,Īnalytical and Quantitative Cytology and Histology 23(4):291-299, 2001. Standard brightfield stain vectors Stain name Measuring theĮmission strength for each dye in each channel again leads to the data to be written in stains to estimateĭye densities using this function. Each dye will be visible in a subset of these channels. In multi-spectral fluorescence imaging, channels are not set up specifically for each dye. Slides prepared for the purpose, with a single dye. light from one dye is partially recorded in a channel set up for a different dye.Īgain, it is possible to measure the emission intensity in each channel (or channel cross-talk ratios) using Typically, fluorescence imaging systems are set up such that each channel collects light only from a single dye,īut in practice it is not always possible to use dyes with perfectly separated emission spectra. Of fluorescence, dip::BeerLambertMapping should not be used. The explanation above translates to fluorescence imaging, replacing ‘absorbance’ with ‘emission’. Functions void dip:: BeerLambertMapping( dip::Image const& in,ĭip::Image::Pixel const& background) Applies a logarithmic mapping to a transmittance image to obtain an absorbance image void dip:: InverseBeerLambertMapping( dip::Image const& in,ĭip::Image::Pixel const& background = ) dip :: Image nuclei = img dip :: Image dab = img Fluorescence Struct dip:: ColocalizationCoefficients Holds Colocalization Coefficients as described by Manders, see dip::MandersColocalizationCoefficients.


 0 kommentar(er)
0 kommentar(er)
